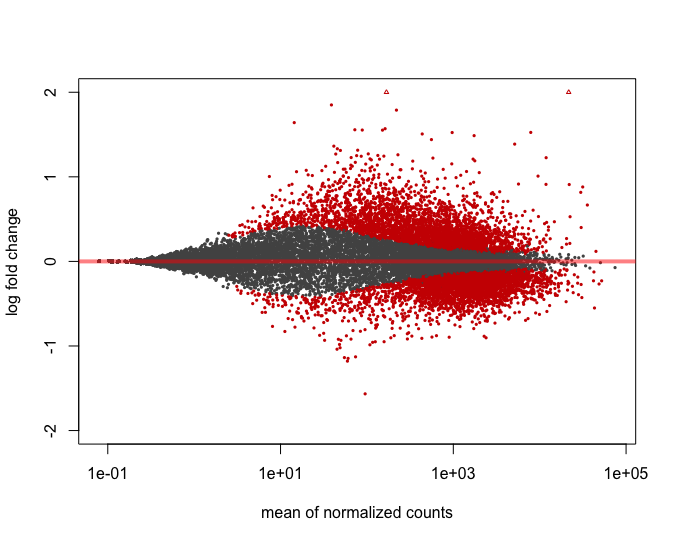
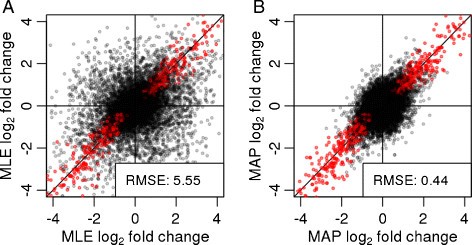
Linear regression fitted between Log2 fold change (FC) in RNA-seq analysis and -ΔΔCt in Biomark qPCR assay of 39 selected DEGs.

Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots | Scientific Reports
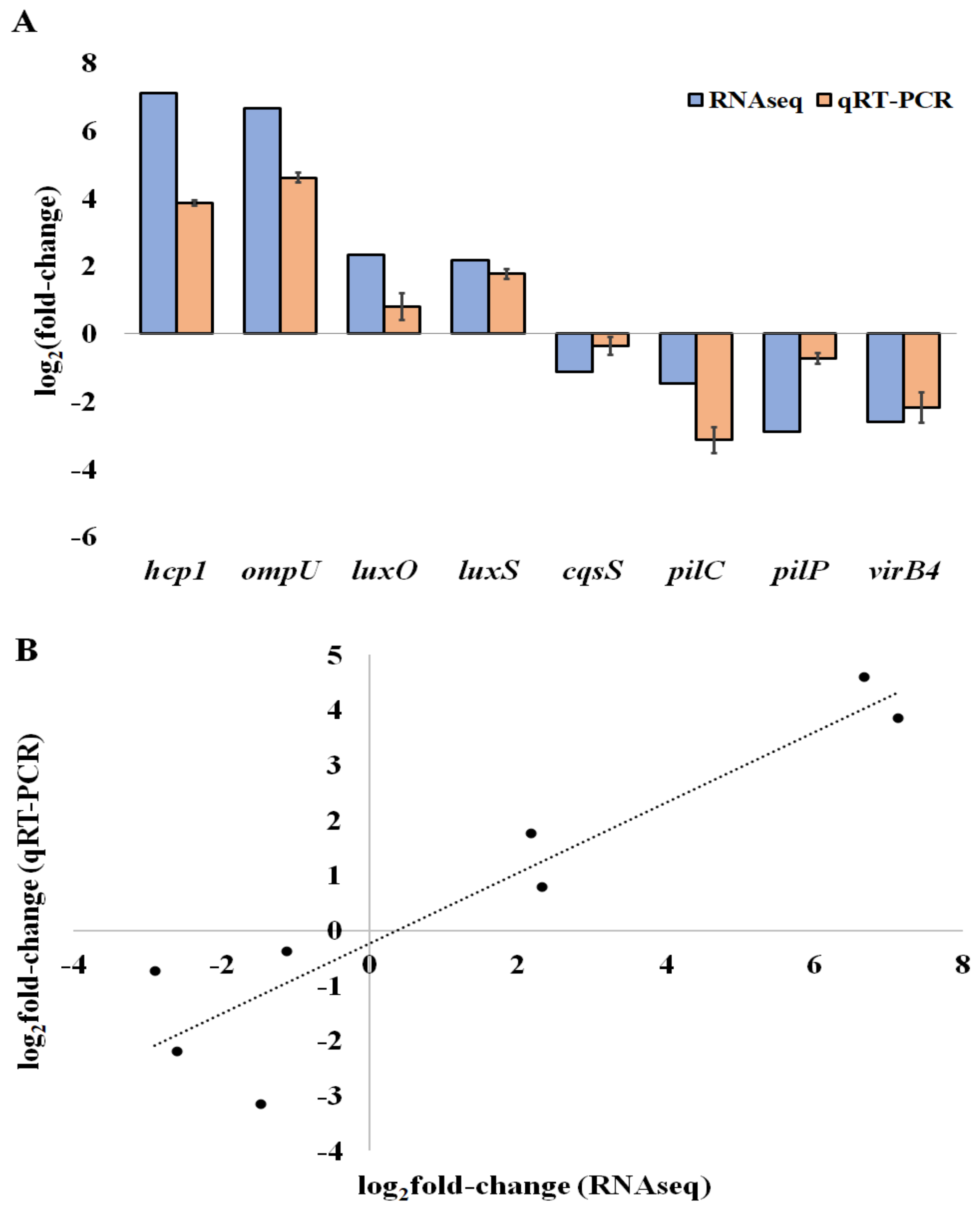
Pathogens | Free Full-Text | Relation between Biofilm and Virulence in Vibrio tapetis: A Transcriptomic Study | HTML